Glikoz update
Check out Glikoz @ GitHub
I have just finished pushing some small changes to the main glikoz branch, and finally I can call it something I’d be okay with using for the rest of my life. This opens up space to doing cool things and making things look better, but first I’ll get into some detail about what are the most important features in glikoz, at least for me.
What we have
First of all, glikoz works with exports from a very well-established OSS app for recording blood sugar and other relevant information, namely Diaguard. This was very important for me from the beginning, coming from proprietary software that was very restrictive in letting users access their own data in a raw format.
With that data, we have all the basic analyses. It is always important for a doctor and their diabetic patients to keep track of how their Time in Range and HbA1c are going. Glikoz reports both the time in range and the estimated HbA1c (you can find references on how the HbA1c is estimated in the GitHub repository). Besides that, the usage of test strips, lancets and fast-acting insulin is reported to help with managing the use of these very expensive resources.
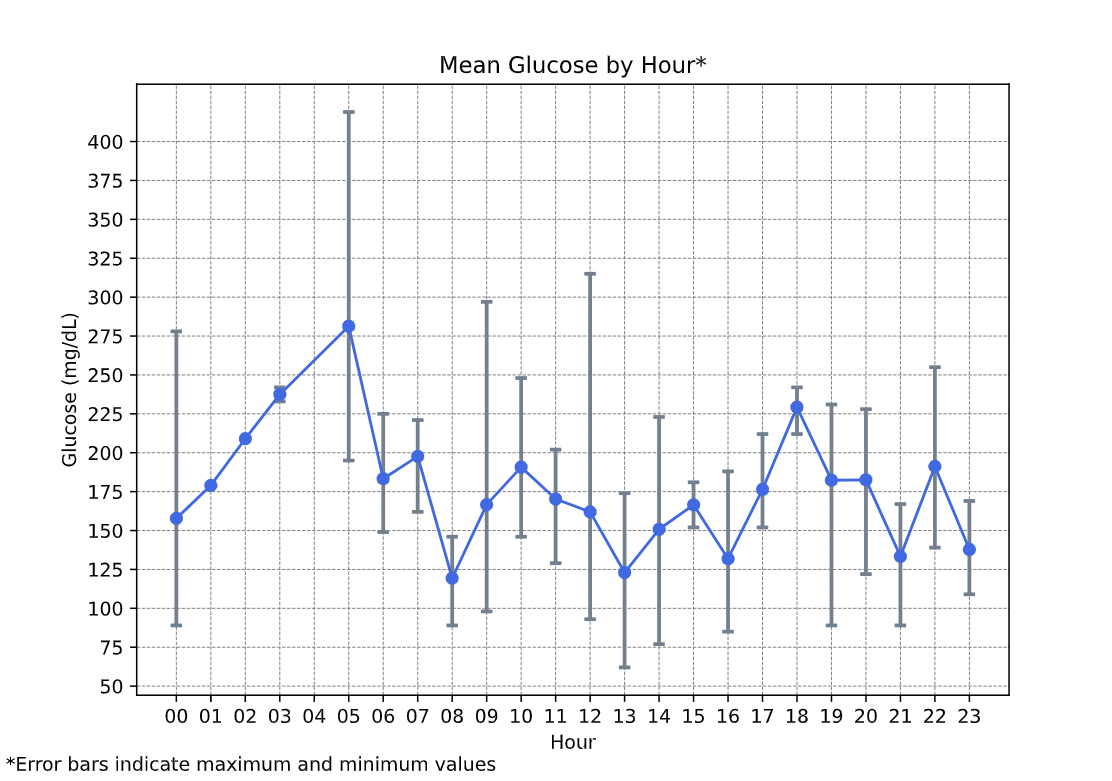
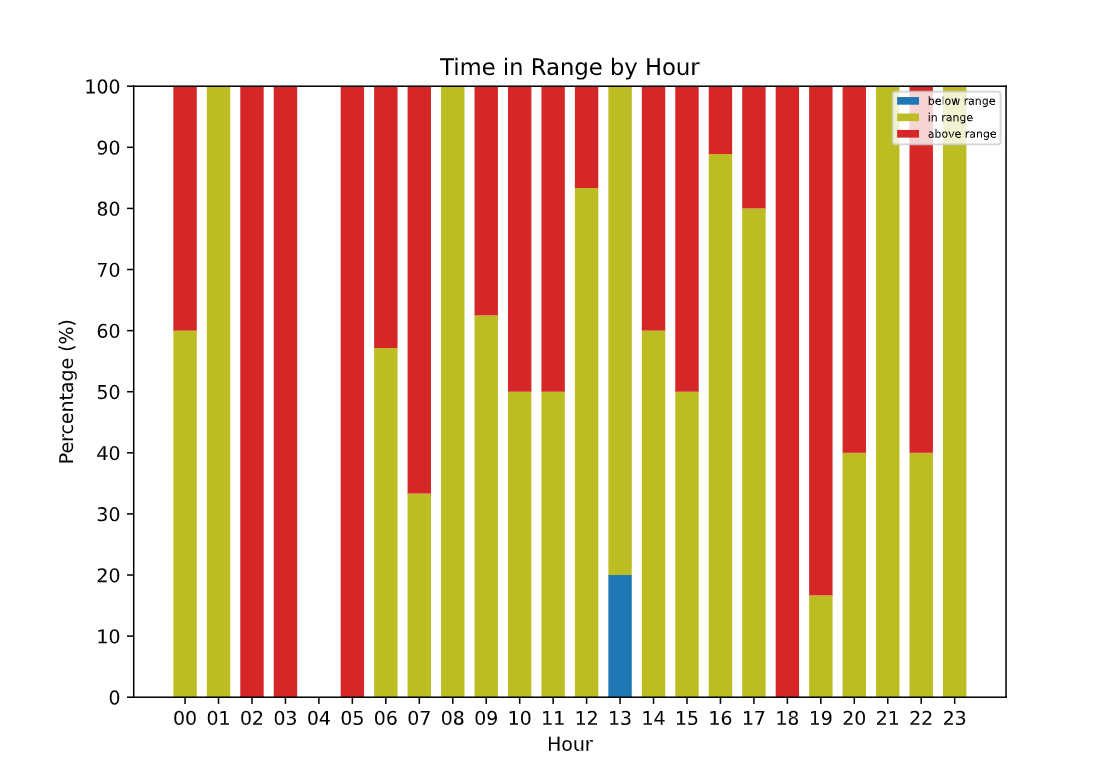
There is also a 24 hour graph with the mean glucose by hour and the reported minimum/maximum as well, and another that presents the Time in Range for each hour instead of all the entries. These are both very important for a less-biased, in-depth review of blood sugar behaviour.
For the PDF reports to be useful in actual day-to-day patient data analysis, glikoz also includes a day-by-day table view of the most recent entries. This allows for a close look at what the treatment is looking like on a daily basis.
Finally, I added a special page for hypoglycemia-related statistics, reporting the number and percentage of episodes and a graphical view of the distribution of episodes among danger levels. Hypoglycemia is one of the most dangerous aspects of Type 1 Diabetes, and I feel other softwares don’t go far beyond the simple time-in-range graph. I would say this is one of the most important features in glikoz right now.
How it was built
I used Python3 for all of glikoz. Since the scope of processing is still very small, and to use it as a proof of concept, Pandas and NumPy are used for data analysis. If you consider that the average user will input about 15 entries every day, we are talking about less than 6 thousand entries (i.e., table rows) in a year, meaning we can allow for even very slow algorithms (think cubic) before starting to worry about execution time.
In the future I plan on switching to something distributed, of course, but Pandas will be more than enough in the meantime.
Plans for the future
There are, of course, infinite features I want to add to glikoz. One of those that will have to wait a while longer, for example, is reporting analyses with Boukeh, which would allow us to create web-based interactive reports. Another is machine learning-powered analysis to provide insights about the treatment and optimization of insulin dosage parameters based on recent data.
The two most important things, though, and these are the ones I’ll be working on next, are documentation and libre integration.
I made sure to write readable code with inline documentation (and since a while ago I’ve been doing test-driven development, which helps with that), but I feel glikoz still lacks a textual explanation of its purpose, architecture and backlog. This is not a coding task, which might be the reason for me not having done it yet, but still it’s very important.
And the Freestyle Libre integration is probably the most useful thing I can do to access more data. This will allow for minute-by-minute analysis of blood sugar behavior. I’m really looking forward to it. Since this means a lot more data, however, I plan on moving to SQL-powered analysis wherever possible. This will also help in scaling later on.
Final remarks
Well, that was it. I truly am excited to continue working on glikoz, and I hope I can learn a lot from these next steps.